In the effective 2-state NMP model a single defect is described by the four parameters: \begin{equation} \boldsymbol{p}=\left(x_\mathrm{T}, E_\mathrm{T}, E_\mathrm{R}, R \right) \end{equation} where \( E_\mathrm{T}\) denotes the energy level of the defect, \( x_\mathrm{T}\) the the position, \(E_\mathrm{R}\) the relaxation energy and \(R\) the curvature ratio. However, most degradation experiments are carried out on large-area devices, so that only the effect of an entire defect ensemble can be observed, not the effect of individual defects. Assuming non-interacting defects, the effective threshold voltage shift \(\Delta V_\mathrm{th} \) caused by a defect ensemble can be expressed as a linear superposition of individual defect contributions \(\delta V_\mathrm{th} \): \begin{equation} \Delta V_\mathrm{th}(t,V_\mathrm{G},T)=\int N(\boldsymbol{p})\cdot \delta V_\mathrm{th}(t,V_\mathrm{G},T;\boldsymbol{p}) \mathrm{d}\boldsymbol{p} \end{equation} with an a priori unknown defect parameter distribution \( N(\boldsymbol{p}) \). Consequently, in order to obtain a well-calibrated model for the degradation of the device, one is faced with the task of inferring the a priori unknown distribution \( N(\boldsymbol{p}) \) of defect parameters from experimental measurements of \(\Delta V_\mathrm{th}\) at accelerated stress conditions. This problem statement can be recast into a non-negative linear least square (NNLS) problem: \begin{equation} N(\boldsymbol{p})=\underset{\hat{N}\geq 0}{\mathrm{arg\,min}} \left|\Delta V_\mathrm{th}-\int_\Omega \hat{N}(\boldsymbol{p})\delta V_\mathrm{th} \mathrm{d}\boldsymbol{p}\,\right|^2\, \end{equation} The naive attempt to solve the optimization problem stated by Eq. 3 would lead to solutions that are very sensitive to noise in the measurement data. To overcome this issue, previous studies often assumed that the defect parameters are distributed according to a Gaussian distribution. This assumption significantly reduces the complexity of the problem since then only the mean and sigma values of the defect parameters need to be optimized to fit the experimental data. However, enforcing an analytic form of \( N(\boldsymbol{p}) \) can lead to unphysical artifacts. In order to circumvent this issue, Comphy offers a novel approach for parameter extraction named Effective Single Defect Decomposition (ESiD), which will be described below.
#n t Vg dVth T
#u s V V K
0.000000000001000 -0.5 nan 425
0.000000001001000 -2.99 nan 425
0.000000010965210 -2.99 nan 425
0.000000120214896 -2.99 nan 425
0.000001318051382 -2.99 nan 425
0.000014451381909 -2.99 nan 425
0.000158447973159 -2.99 nan 425
0.001737256809792 -2.99 nan 425
0.019047648125279 -2.99 nan 425
0.208842410190570 -2.99 nan 425
2.289792000000999 -2.99 nan 425
2.291232001000997 -0.54 -0.003961323371012737 425
2.293856001000998 -0.54 -0.0033636287968579204 425
2.300704001000998 -0.54 -0.0031793755039067895 425
2.321120001000999 -0.54 -0.00256470368748718 425
2.370656001000999 -0.54 -0.001904752848599256 425
2.511072001000999 -0.54 -0.001568746921409958 425
2.906208001000998 -0.54 -0.0014124223875647823 425
3.930368001000998 -0.54 -0.0012746819560973677 425
6.834112001000999 -0.54 -0.00120204805313473 425
14.457536001000999 -0.54 -0.0014834494304144519 425
- The first line must start with #n and contain the names of the columns using the same names as shown above. The second line must start with #u and contain the units. No unit conversion is performed at the moment, but might be added at a later point.
- If no \( \Delta V_\mathrm{th} \) measurement was carried out at a certain point in time, the corresponding entry in the time-stress file must be replaced by "nan". The framework automatically excludes the corresponding point from the fitting process.
- The ESiD algorithm allows for the simultaneous fitting of multiple datasets. For this purpose, simply save the different data sets as separate time-stress files.
The following example shows the typical format of an input file. Each parameter begins with two dashes, followed by the parameter name and the parameter value separated by a space. The first part of the input file lists the necessary device parameters while the second part references the time-stress files containing the measurement data.
--Tinit 300
--Na_chan 1e+10
--Nd_chan 5.5e+17
--Eg0_chan 1.206
--Eg1_chan -2.73e-4
--Eg2_chan 0.0
--Egd_chan 636.0
. . . . . .
--tsFile[0] 'table_pMOS_VgS-2.99_T125.0.crv'
--tsFile[1] 'table_pMOS_VgS-3.888_T125.0.crv'
inputFile =
params, setups = _partofmain([
my_device = V3Device(**params).device
| |
|
| |
|
| |
|
| |
|
| |
|
| |
|
| |
|
| |
|
| |
|
| |
|
| |
|
| |
|
| |
|
| |
|
| |
|
| |
|
| |
|
| |
|
| |
|
| |
|
| |
|
E.optimize()
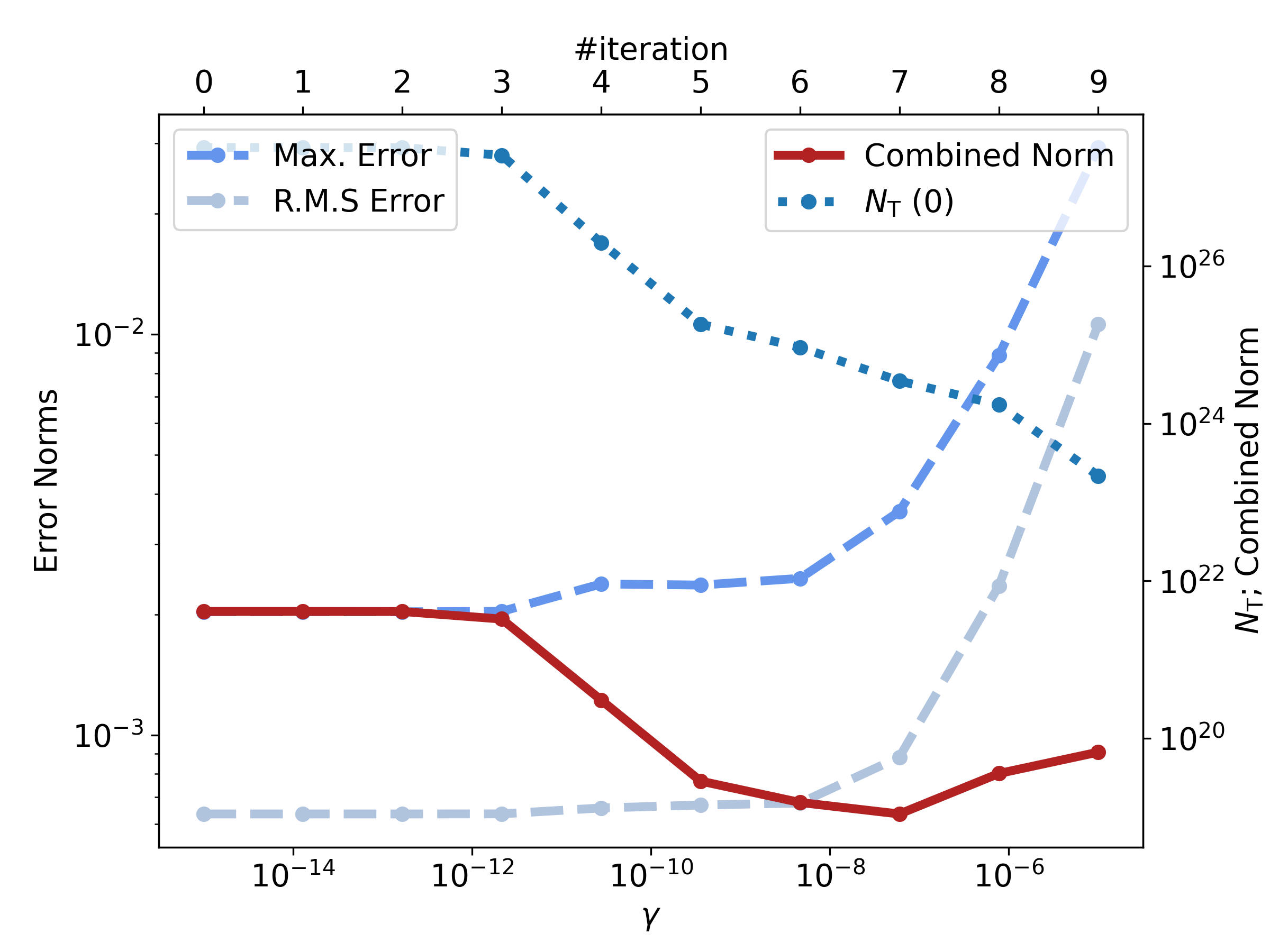
INFO: Iteration Number = 00
INFO: Gamma = 1.000e-09
INFO: Max. Error = 2.582e-04 V
INFO: R.M.S Error = 9.977e-05 V
INFO: Estimated Nt(0) = 2.178e+27 m⁻³
INFO: Combined Norm = 5.612e+19 V²m⁻³
INFO: output data written to path ./Results/ESiD_run_1/00
INFO: #########################################
INFO: #########################################
INFO: Iteration Number = 01
INFO: Gamma = 3.162e-09
INFO: Max. Error = 2.597e-04 V
INFO: R.M.S Error = 1.026e-04 V
INFO: Estimated Nt(0) = 5.386e+26 m⁻³
INFO: Combined Norm = 1.436e+19 V²m⁻³
INFO: output data written to path ./Results/ESiD_run_1/01
INFO: #########################################
. . . .
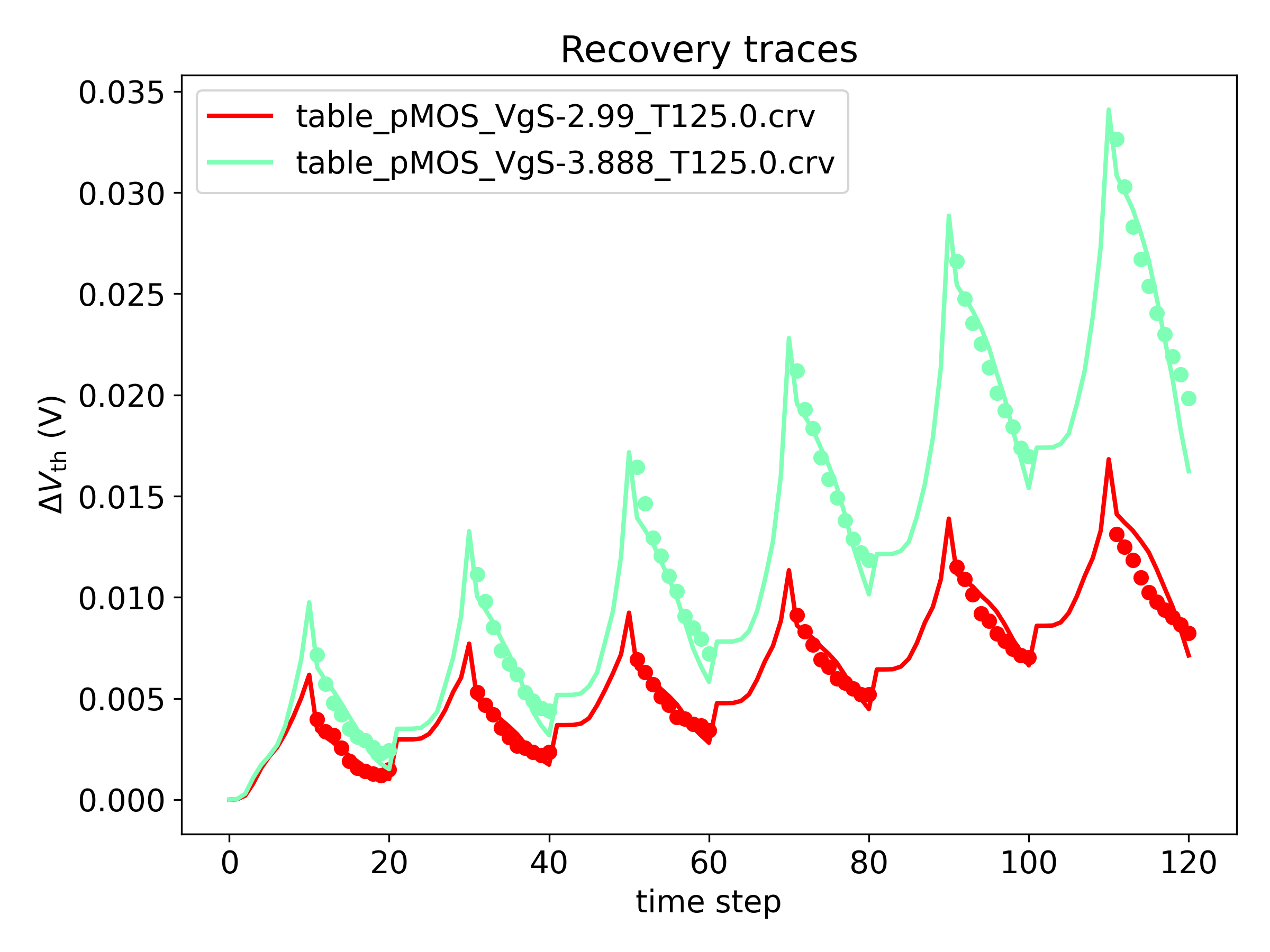
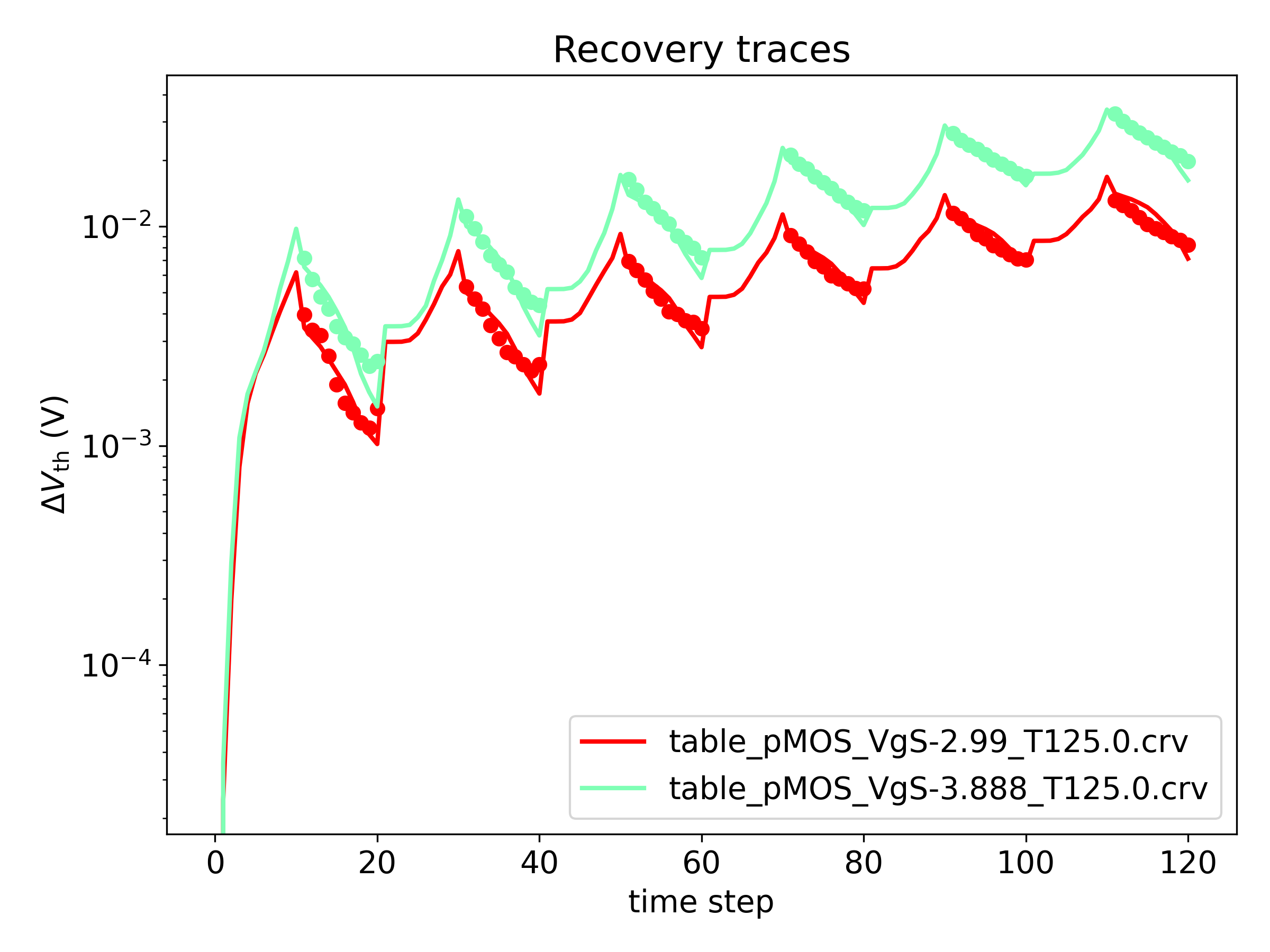
Plot of measured and fitted \( \Delta V_\mathrm{th} \)
- The plot 'linear_msm.png' shows the mentioned comparison with a linear y-scale:
- The plot 'log_msm.png' shows the above comparison on a logarithmic y-scale:
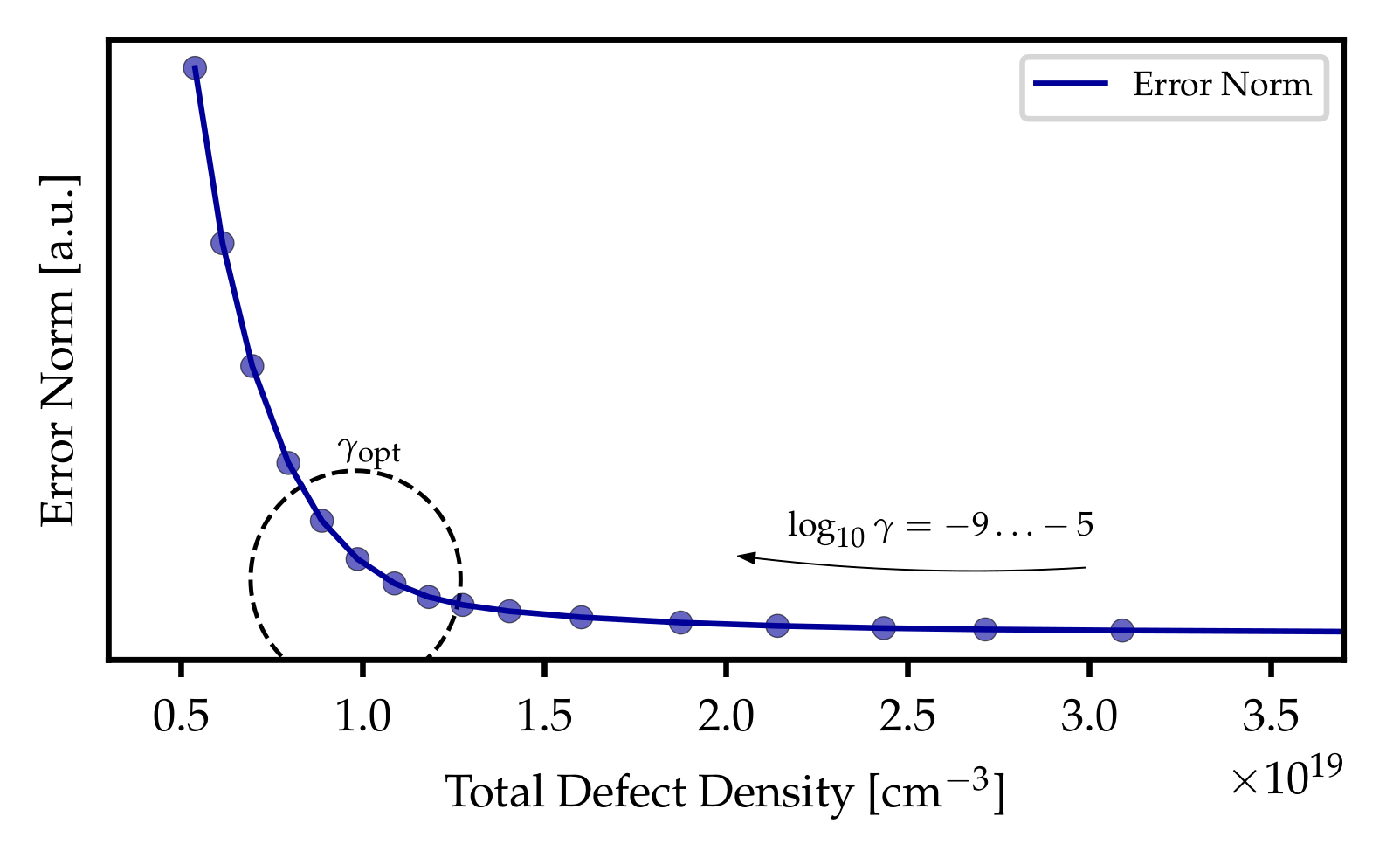
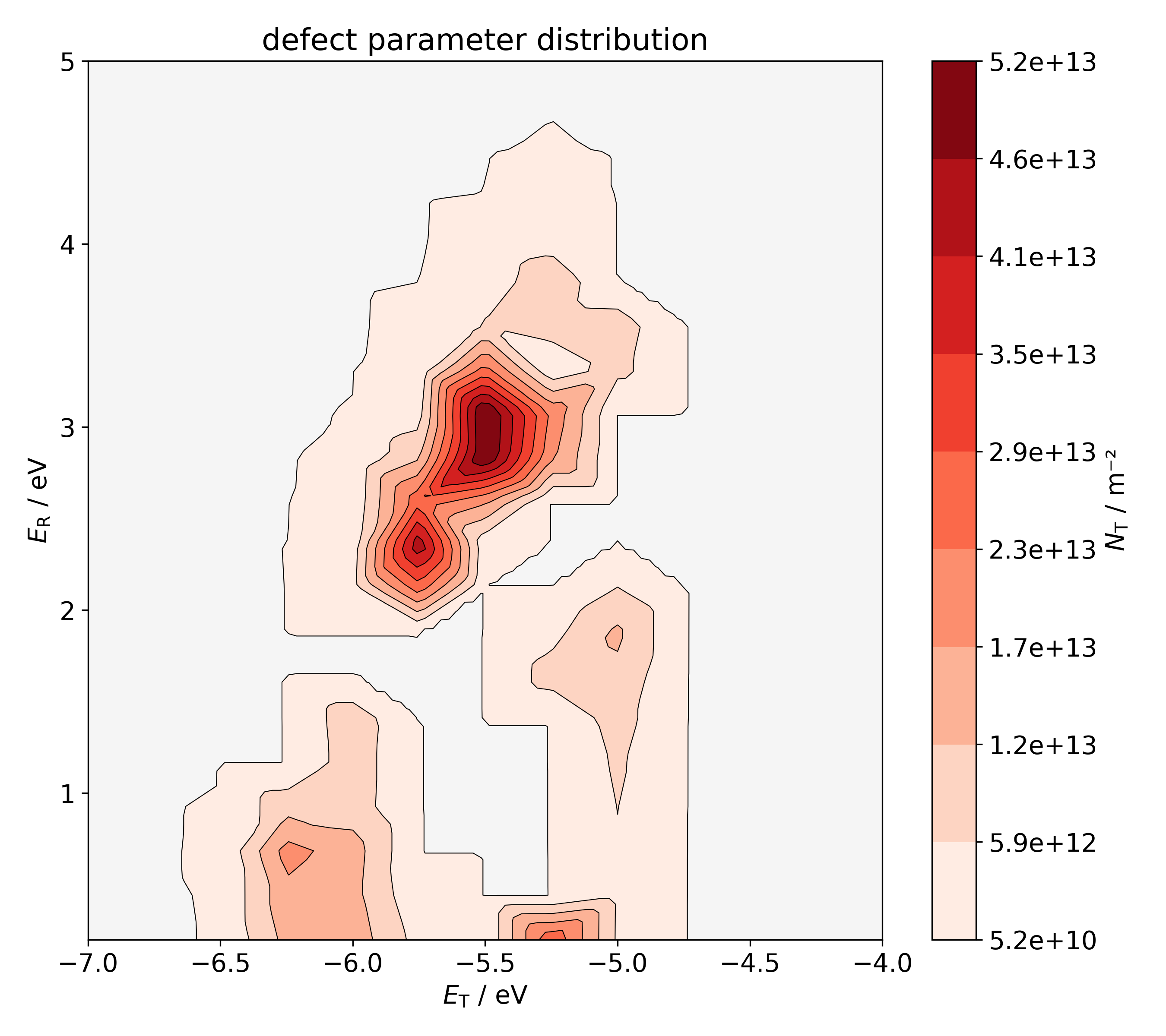
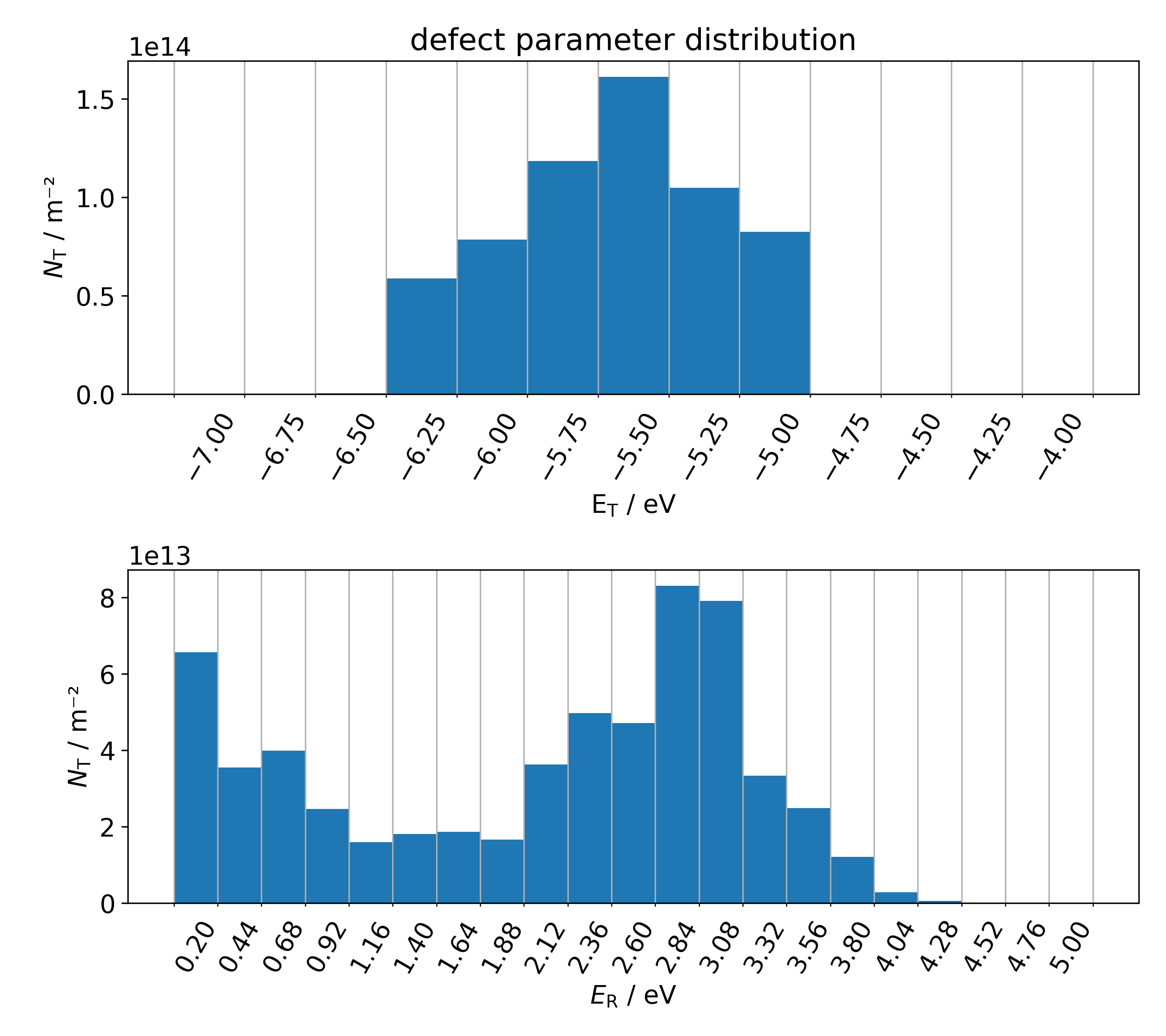
Fitted defect parameter distibutions
- The plot 'i_2Dcont.png' shows a continuous contour plot of the defect parameter distribution \( N( E_\mathrm{T}, E_\mathrm{R}) \) of the ith trap band. Since the defect densities are only known on the discrete grid used for the optimization, linear interpolation is applied to construct this continuous plot:
- In addition, the file 'i_1Dhist.png' shows the one-dimensional projections of the defect parameter distribution \( N( E_\mathrm{T}, E_\mathrm{R}) \) of the ith trap band:
time-stress file containing the measured and fitted \( \Delta V_\mathrm{th} \)
# Version: 3.0
#n ts Vg dVthSim T dVthMeas
0.000000000 -0.500000000 0.000000000 425.000 nan
0.000000001 -2.990000000 -0.000103815 425.000 nan
0.000000011 -2.990000000 -0.000893064 425.000 nan
0.000000120 -2.990000000 -0.002742621 425.000 nan
0.000001318 -2.990000000 -0.004521839 425.000 nan
0.000014451 -2.990000000 -0.006204676 425.000 nan
0.000158448 -2.990000000 -0.008219382 425.000 nan
0.001737257 -2.990000000 -0.009891099 425.000 nan
0.019047648 -2.990000000 -0.010742648 425.000 nan
0.208842410 -2.990000000 -0.011003577 425.000 nan
2.289792000 -2.990000000 -0.012035140 425.000 nan
2.291232001 -0.540000000 -0.003731345 425.000 -0.003961323
2.293856001 -0.540000000 -0.003027215 425.000 -0.003363629
2.300704001 -0.540000000 -0.002487522 425.000 -0.003179376
2.321120001 -0.540000000 -0.002163318 425.000 -0.002564704
2.370656001 -0.540000000 -0.001966182 425.000 -0.001904753
2.511072001 -0.540000000 -0.001879344 425.000 -0.001568747
2.906208001 -0.540000000 -0.001831552 425.000 -0.001412422
3.930368001 -0.540000000 -0.001800012 425.000 -0.001274682
6.834112001 -0.540000000 -0.001752228 425.000 -0.001202048
. . . .
error plot